- About
- Organization
- Organization Overview
- Dean’s Office
- Department of Bioengineering and Therapeutic Sciences
- Department of Clinical Pharmacy
- Department of Pharmaceutical Chemistry
- Quantitative Biosciences Institute
- Org Chart
- Research
- Education
- Patient Care
- People
- News
- Events
Taking a bite out of the proteome with PhaNGS
New technology promises to speed the study of protein biology in health and disease
By Levi Gadye / Wed Mar 7, 2018
If DNA is the blueprint for every cell in the body, then proteins are the cell’s construction workers, forklifts, and building materials. However, unlike DNA, which can be readily sequenced, proteins are difficult to sequence and identify, creating a formidable hurdle to understanding how cells work or how proteins misbehave during diseases like cancer.
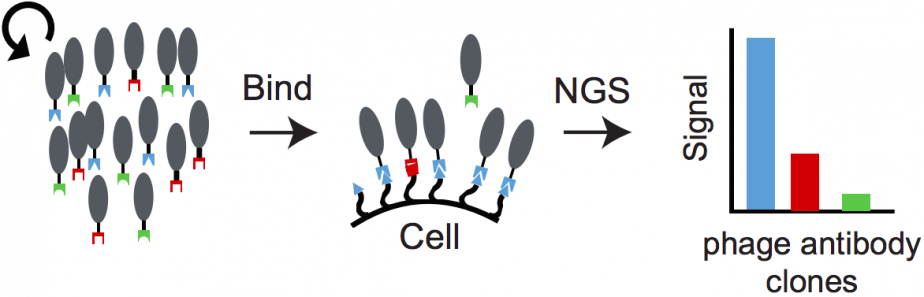
Schematic of PhaNGS method. From left: a library of phages that bind to proteins of interest is assembled; a cell is exposed to this library, and unbound phages are washed away; bound phages are identified and their relative levels are measured using NGS (from Figure 1B, Pollock et al., PNAS 2018).
Recent work in the UCSF School of Pharmacy could make the proteome—the word for the sum of all of a cell’s proteins—nearly as readable as the genome itself. The research, published online February 23 in PNAS8, uses viruses to flag individual proteins on the surface of cells with a unique “DNA barcode,” which can then be read using next-generation sequencing (NGS). The new method, developed by a team led by James Wells, PhD9, is called “phage antibody next generation sequencing,” or PhaNGS.
Traditionally, to study proteins, scientists had to start by knowing exactly which protein they were looking for. “You can follow that protein around in its function, or during its birth and destruction, [by using antibodies],” says Wells, a faculty member in the School’s Department of Pharmaceutical Chemistry10 and a member of the Helen Diller Family Comprehensive Cancer Center11 at UCSF. Wells is the senior author on the paper.
Antibodies, which are proteins themselves, are designed to attach to a specific target protein according to their unique, complementary shapes, much like a lock and a key. Unfortunately, commercial antibodies are expensive, and even the best methods can only use handfuls of different antibodies at the same time.
More recently, scientists have used mass spectrometry to look at many proteins at once, rather than one protein at a time. Mass spectrometry breaks up a group of proteins into small pieces. Scientists can laboriously reconstruct proteins from the smaller pieces, as if they were a microscopic 3D puzzle. The process gives a bird’s eye view of hundreds of proteins at once, but it has its limitations—and a steep price tag.
Mass spectrometry “requires huge numbers of cells, it's expensive, and it merely detects fragments of proteins,” making it challenging to study the shapes of entire proteins, which often determine their functions, says Wells.
Wells' group, along with collaborators at the University of Toronto, wanted to identify intact proteins without the constraints of either traditional antibody-based methods or mass spectrometry. They turned to phages—viruses that infect bacteria by sticking to proteins on the bacterial cell membrane. This process is analogous to how antibodies stick to target proteins; but, unlike traditional antibodies, phages also contain DNA that determines what they stick to. This DNA encodes for the phage’s outer, sticky “fingers,” and, crucially, this DNA can be easily deciphered using NGS.
Given the importance of the proteome to nearly every aspect of biology and human health, PhaNGS holds immense promise for the biomedical sciences.
A few years ago, Wells' team wanted to design phages to stick to proteins of interest, rather than bacteria, so they replaced one of the sticky fingers with a fragment of a known antibody (and cancer drug), Trastuzumab. Then, this antibody fragment was systematically mutated, resulting in 10 billion unique phages. Each mutant phage possessed a unique antibody fragment—and with luck, some might stick to cancer-associated proteins.
The group of phages was then exposed to individual cancer cell markers, and any phages that didn’t stick were washed away. Of the 10 billion phages, nearly 200 were identified that stuck to about 50 cancer proteins. Normally, antibodies that precisely stick to known targets must be generated in animals, but Wells’ team had accomplished this using viruses in test tubes, at a fraction of the cost.
Taking this method a step further, the team recently surmised that they could easily assess which cancer proteins were present on the surface of cells, and in what proportions, using NGS to track the presence of each of the 200 phages at the same time. First author and graduate student Sam Pollock tested the 200 phages on tissue samples taken from a cancer patient, before and after chemotherapy, as well as on a cell culture model of cancer, and then measured the amount of each phage that remained on the cells, using NGS. In each case, the method revealed how the assortment of cell-surface proteins changed either in response to chemotherapy or the onset of cancerous growth. Since the method is paired with sequencing, the team coined the term “phage antibody next generation sequencing,” or PhaNGS.
But the true value of PhaNGS goes beyond cancer biology. “It's way cheaper than mass spectrometry and it detects protein conformations (or shapes), which mass spectrometry cannot do,” says Wells.
Importantly, PhaNGS also works on individual cells—a boon to biologists, who increasingly turn to single-cell methods to study different cell types, like the cells that seed the growth and metastasis of tumors.
Wells doesn’t think PhaNGS will completely replace mass spectrometry, but is hopeful the technology will give scientists a more sensitive tool for fully understanding the proteome. “These are complementary methods,” he says. “Mass spectrometry doesn't require any antibodies at all, and it doesn't require phage libraries. But it doesn't tell you anything about the 3D structure of the protein that was there, while antibodies—and PhaNGS—can detect that.”
The library is poised to swell even further as PhaNGS is applied to other scientific questions. So far, a dozen collaborators have lined up to begin using the technology in their own labs.
Given the importance of the proteome to nearly every aspect of biology and human health, PhaNGS holds immense promise for the biomedical sciences. Wells now has set his sights on developing phages that target all the proteins on the cell surface—proteins that are vital for determining a cell’s health and behavior. “It's a growing library, and we want to have antibodies to every single cell surface protein,” Wells says, “all 4,000 of them.”
Additional authors on the study include Amy Hu, Yun Mou, Alexander J. Martinko, Olivier Julien, Michael Hornsby, Lynda Ploder, Jarrett J. Adams, Huimin Geng, Markus Müschen, Sachdev Sidhu, and Jason Moffat.
Tags
Keywords:
Category:
Research18
Sites:
School of Pharmacy, Department of Pharmaceutical Chemistry, PharmD Degree Program, CCB, PSPG, Biophysics
About the School: The UCSF School of Pharmacy aims to solve the most pressing health care problems and strives to ensure that each patient receives the safest, most effective treatments. Our discoveries seed the development of novel therapies, and our researchers consistently lead the nation in NIH funding. The School’s doctor of pharmacy (PharmD) degree program, with its unique emphasis on scientific thinking, prepares students to be critical thinkers and leaders in their field.





